Architecture
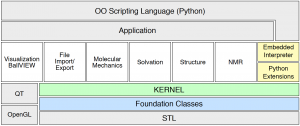
The overall structure of BALL may be seen as consisting of several layers, each providing functionality for a well-defined area. In this layered architecture the layers higher up depend on the lower ones for their implementation.
The lowest layer contains the so-called Foundation Classes. These classes implement a set of general data structures like an extended string class, hash maps, or design patterns. The implementation of this layer is based on the standard template library.
The second layer consists of the kernel classes, implementing data structures for atoms, molecules, and the like.
The third layer consists of several basic components. Each of these components provides functionality for a clearly defined field of molecular modeling and is not physically dependent on any of the other basic components (well, at least not very much dependent :). The basic components are implemented using the kernel classes and the foundation classes. They provide support for file I/O in different formats, molecular mechanics, three-dimensional visualizaion, search for structural similarities, for the calculation of nuclear magnetic resonance (NMR) spectra, and solvation methods.
The visualization component relies on OpenGL for platform-independent 3D graphics and on QT for a portable graphical user interface (GUI).
BALL classes can also be used as extensions in the object-oriented scripting language Python and it is possible to embed this scripting language into BALL applications. Besides the rapid prototyping capabilities of the library itself, this provides a very efficient method to create software prototypes and improves the capabilities of BALL applications through the embedding of a scripting language.
 Web Design MymensinghPremium WordPress ThemesWeb Development
Web Design MymensinghPremium WordPress ThemesWeb Development 
